Sage Dynamics Demo
Intro
[This is an adaptation of a page from my open lab notebook wiki, as a demo of our Working Markup reproducible web-publishing system. Scroll down to the final section for info on how this post is made, including source code. -lw]
This post demonstrates a set of Sage classes I wrote to work with continuous-time dynamical systems, the kind of system my colleagues and I use a lot as mathematical models.
The computations in this blog post are implemented in two parts. The Sage class library is kept in a GitHub repository by itself, and the Sage scripts that use the library to implement a simple demo system on this page are stored as part of this page.
On this page I use those Sage classes to construct a simple population biology model of two populations, and analyze and evaluate it.
The Sage code for this model is below. Here’s its output, in LaTeX format:
The generic competition model:
The competition model with parameters bound to specific values:
Equilibria of the generic model:
Stable equilibria of the bound model:
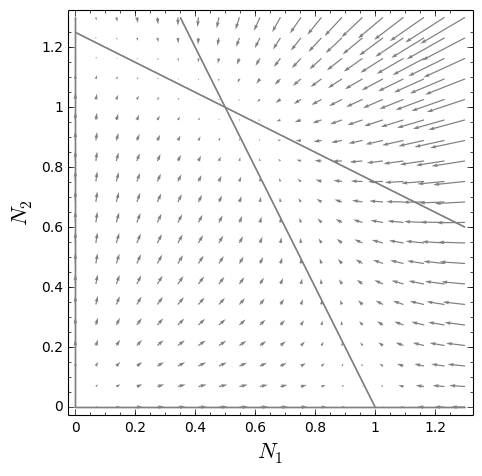
Here’s the state space plot that the program creates, showing a representative trajectory of the model’s dynamics, with flow vector field and zero-net-growth isoclines:
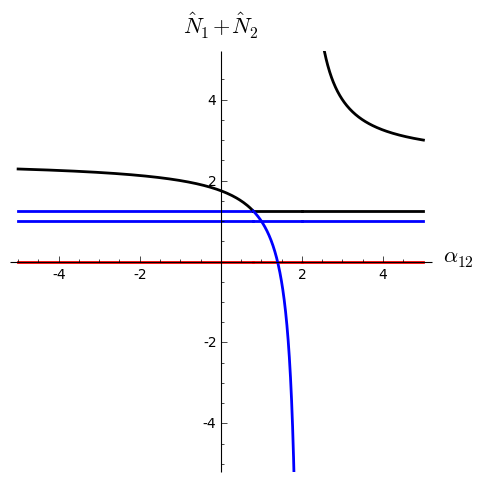
And a bifurcation diagram of total population vs. (black=stable, red=repelling point, blue=saddle):
Source code
Here is the source code for this page’s demo:
And a small makefile to get the above Sage code to connect with the libraries pulled from GitHub into a separate directory:
Reproducibility
Here are instructions to run this Sage code yourself. You will need Sage, GNU make, git, perl, and maybe some other utilities installed.
- Download and unpack the source code from this page: wmd_export.tgz
- Install Sage and the Sage library code from https://github.com/worden-lee/SageDynamics
- In the code directory, run
make ode-system-demo.sage.out.tex
To see how this blog post is written, with source code embedded, see https://github.com/worden-lee/github-pages-sandbox/blob/master/_posts/2014-10-20-SageDynamics.markdown.wmd.